- Home
- About
- OSD
- MyOSD
- Partners
- Work Packages
- WP 1Management & Coordination
- WP 2 (OSD)Ocean Sampling Day
- WP 3Oceanography & Environmental Data
- WP 4Standards and Interoperability
- WP 5Bioinformatics & Data Integration
- WP 6Exploring Ecosystems Biology
- WP 7Function and Biotechnology
- WP 8Intellectual Property (IP) Management for Marine Bioprospecting
- WP 9Dissemination & Outreach
- Public DeliverablesAll Micro B3's public deliverables
- Meetings
- Workshops
- Third Micro B3 Industry Expert Workshop
- Micro B3 Industry Expert Workshop
- Micro B3/OSD Analysis Workshop
- Micro B3 Stakeholder Workshop
- Micro B3 Summer School in Crete 2014
- Marine Metagenomics Bioinformatics
- Micro B3 Industry expert workshop
- EU-US Training 2013
- Micro B3 Statistics Training 2013
- MG4U Bioinformatics Training 2013
- Bioinformatics Training 2012
- EU-US Training 2012
A new axis of collaboration: BioVeL workflows for the Micro B3 Ocean Sampling Day
A new axis of collaboration: BioVeL workflows for the Micro B3 Ocean Sampling Day
On March 21 and 22, partners from Micro B3 and the FP7 EU project Biodiversity virtual e-Laboratory (BioVeL) met in Bremen. Altogether, 16 scientists and engineers worked on designing workflows for the Ocean Sampling Day (OSD) and create a plan for implementing these workflows based on the web services of the e-infrastructure of BioVeL.
In the introduction to the meeting an update on the status of the Micro B3 developments and the scientific goals of OSD was presented. This was followed by a series of talks on introducing BioVeL and giving a status report on the e-infrastructure components already in place.
In an open discussion plans for five prime workflows, of use to OSD, were developed (Figure 1):
For single OSD samples:
- Microbial Community Phylogenetics
- My Microbial Community
For comparative analysis:
- Microbial Assemblage Prediction
- Metagenomic Traits Analysis
- Single Gene Cluster Analysis
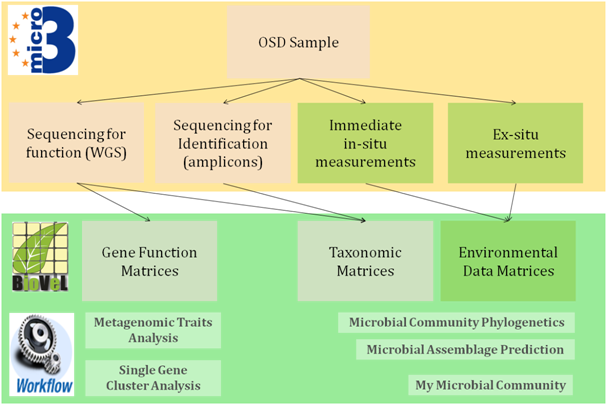
Figure 1: The Micro B3/OSD data flow and BioVel workflows
All workflows will be made available to the OSD and also wider community for consistent and repeatable metagenomic sequence data analysis and microbial community modelling. Moreover, using workflows the data analysis will be more transparent and the provenance automatically recorded. Every OSD participant can make use of these workflows, rendering the analysis of the Ocean Sampling Day data a community effort.
With the majority of OSD samples being amplicon based metagenomics for taxonomic identification (tags, barcodes), the current focus of the developments lies on supporting the analysis of this kind of data. These workflows have a common set of pre-requisites: they all need already pre-calculated data matrices (Figure 1), which will be delivered by the Micro B3 Information System. It became also apparent, that a number of BioVeL web services and workflow components from metagenomics, phylogenetics and ecological niche modelling can be re-used for the new OSD workflows, with only a few additional services to be developed.
Forming a unique axis of collaboration, BioVeL together with Micro B3 is now developing e-science workflows, that will be made available to the OSD community for consistent and repeatable metagenomic sequence data analysis and microbial community modelling. Each individual OSD participant will benefit from an infrastructure for consistent and repeatable analyses of their own microbial sequence data; whereas BioVeL can broaden its portfolio of workflows and services supported by Micro B3 scientists, delivering products that will demonstrate the usability of its e-science infrastructure to the new Micro B3/OSD scientific community. This workshop kick-started developments which will provide the two communities with a wealth of data and unique insights into the microbial mechanisms underpinning the world’s oceans.
Figure 2: From left to Right: (back row) Alan Williams, Frank Oliver Glöckner, Alex Hardisty, Julia Schnetzer, Katja Lehmann, Bachir Balech, Anna Klindworth, Saverio Vicario, Bruno Fosso, (front row) Norman Morrison, Antonio Fernandez Guerra, Dawn Field, Mesude Bicak, Renzo Kottmann, Giacinto Donvito, Stian Soiland-Reyes.
Figure 3: Discussing the workflows for Micro B3/OSD




