- Home
- About
- OSD
- MyOSD
- Partners
- Work Packages
- WP 1Management & Coordination
- WP 2 (OSD)Ocean Sampling Day
- WP 3Oceanography & Environmental Data
- WP 4Standards and Interoperability
- WP 5Bioinformatics & Data Integration
- WP 6Exploring Ecosystems Biology
- WP 7Function and Biotechnology
- WP 8Intellectual Property (IP) Management for Marine Bioprospecting
- WP 9Dissemination & Outreach
- Public DeliverablesAll Micro B3's public deliverables
- Meetings
- Workshops
- Third Micro B3 Industry Expert Workshop
- Micro B3 Industry Expert Workshop
- Micro B3/OSD Analysis Workshop
- Micro B3 Stakeholder Workshop
- Micro B3 Summer School in Crete 2014
- Marine Metagenomics Bioinformatics
- Micro B3 Industry expert workshop
- EU-US Training 2013
- Micro B3 Statistics Training 2013
- MG4U Bioinformatics Training 2013
- Bioinformatics Training 2012
- EU-US Training 2012
Function and Biotechnology
Rationale and objectives
 The objectives of work package 7 (WP 7) are to develop, validate, and apply bioinformatics-based tools and laboratory-screening protocols for the discovery, description, and exploitation of new gene functions. These are enzymatic activities and biosynthetic pathways of bioactive compounds, which can be applied in industrial and pharmaceutical biotechnology. The discovery of novel compounds with biological activity is extremely important since it provides leads for development of biological protection agents, as well as compounds that may be further developed into, for example, antibiotics, food preservatives, or cytotoxic agents with pharmacological potential.
The objectives of work package 7 (WP 7) are to develop, validate, and apply bioinformatics-based tools and laboratory-screening protocols for the discovery, description, and exploitation of new gene functions. These are enzymatic activities and biosynthetic pathways of bioactive compounds, which can be applied in industrial and pharmaceutical biotechnology. The discovery of novel compounds with biological activity is extremely important since it provides leads for development of biological protection agents, as well as compounds that may be further developed into, for example, antibiotics, food preservatives, or cytotoxic agents with pharmacological potential.
The WP 7 team consists of academic and private-sector partners within Micro B3. It uses different approaches:
- Bioinformatics approaches provide tools to establish the function of orphan genes in metabolic pathways and biosynthetic routes. In case of sequence homology to known genes, multiple sequence alignment, homology modelling, and active-site inspection by molecular docking are used to generate hypotheses on the function of specific genes.
- Experimental approaches then explore these bioinformatics-based predictions on the function of genes using whole genome sequences, metabolic maps, and genetic manipulation tools. Cultivation-dependent as well as approaches based on metagenomic libraries are utilized. In the latter DNA is isolated and expressed in a host strain or sequenced, without isolation of pure cultures. Both approaches aim at a broader host range than only E. coli, which is important because expression of some potentially useful activities may be dependent on the host.
One of the challenges of the WP 7 is to access rare members of specialized communities and their unique bioactivities. The discovery of these rare microorganisms may require enrichments.
Recent Progress:
Progress in bioinformatics was achieved by MPI Bremen establishing a system to identify networks of co-occurring unknown genes. The network is constructed using sequences of a number of marine sites. If, within the network, correlations to genes with known functions are found, this hints at a hypothetical function of the unknown genes.
Partner Bio-Prodict, an SME, has established new databases and pursued a further bioinformatic approach in close cooperation with the University of Groningen (UGRO). The databases include data on enzymes like epoxide hydrolases; CoA ligases and on those domains of non-ribosomal peptide synthetases, which play a role in the activation of amino acids for the biosynthesis of secondary metabolites. The University of Groningen is exploring computational approaches, especially high-throughput docking simulations, based on homology models for predicting enzyme activity from sequence.
The industrial partner PharmaMar is focused on genome mining for specific sequences that are part of gene clusters encoding enzymes for the biosynthesis of antitumor compounds.
Left: Using the approach developed at the MPIMM any sequenced genome can be extracted from the co-ocurring network of unknowns and explore the associations within the annotated genes and the hypothetical proteins providing a valuable tool for functional hypothesis generation. In the graph the Pelagibacter ubique HTCC1062 genomic network with an example of how hypothetical proteins can be put biological context is shown.
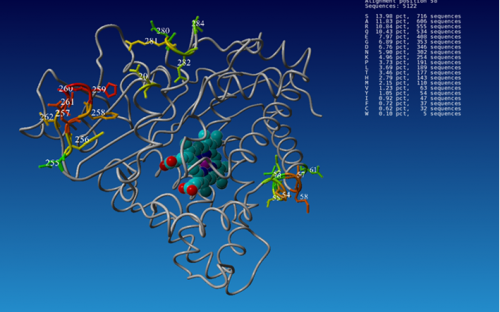
Right: The graph shows a 3D-visualisation of natural amino acid distribution at flexible spots in a cytochrome P450 enzyme using 3DM/Yasara. The natural diversity at flexible positions is displayed in the structure. This information is needed to predict which substitutions are likely to produce an active enzyme when mutated and provides guidance to structure-based mutation design (Bio-Prodict).
For experimental screening, two approaches are followed:
- For screening with cultivation-dependent methods, a range of microbial cultures has been isolated by the Icelandic government-owned company MATIS and other partners:
- IAMC, a national research institute from Sicily, and MATIS have isolated new strains belonging to at least five genera at different salinity and temperature from extreme sites for example collected from Mediterranean deep sea locations and from a coastal hydrothermal vent.
- Based on results of high-throughput screening for cytotoxicity, PharmaMar is selecting organisms that produce interesting bioactive compounds.
- Further high-throughput work is performed on microbial cultures from sponges by BIOMERIT at the University of Cork aimed at the production of enzymes, quorum-sensing compounds, and anti-bacterial compounds.
- For screening with cultivation-independent methods:
- Metagenomic DNA collected at the deep anoxic brine lake Kryos in the Mediterranean has been sequenced, aligned and annotated.
- MATIS had constructed fosmid libraries with over 3,000 clones from in situ enrichments based on tidally influenced hot springs (4-75°C) for several weeks.
Further screening assays are now available and applied to newly created libraries from marine samples
- at the University of Groningen, for discovery of enzymes involved in the deamination of organic nitrogen compounds;
- at the University of Bangor, using various chromogenic plate assays are used;
- at BIO-ILIBERIS, a Spanish SME, for the transformation of xenobiotics compounds by oxidative reactions.
Using these methods several hits have been obtained and work on the characterization of the encoded enzymes is ongoing. This will yield a toolbox of enzymes that may be further evaluated by internal and external parties.
Lead of WP 7: Dick Janssen, University of Groningen